The proximate and ultimate causes of behavioral evolution
The behaviors of closely related species can be remarkably different, and these differences can have important biological consequences. For example, behaviors can be critical to the creation and maintenance of biodiversity, as host, habitat, and mating preference behaviors are often key players in speciation and local adaptation. Despite the importance of behavioral traits, we know little about the genetic and neurological basis of their evolution. This means we have little understanding of the number, type, or cellular effects of mutations resulting in behavioral divergence. These data are important to understanding the tempo and mode of behavioral evolution. To fill this gap, my research seeks to answer the following: (1) what are the genetic loci of behavioral divergence? (2) how do genetic changes affect nervous system structure or function? and (3) how do the selective pressures that drive behavioral evolution shape genetic and neuronal architecture?
Current projects
The genetic basis of behavioral plasticity and its evolution
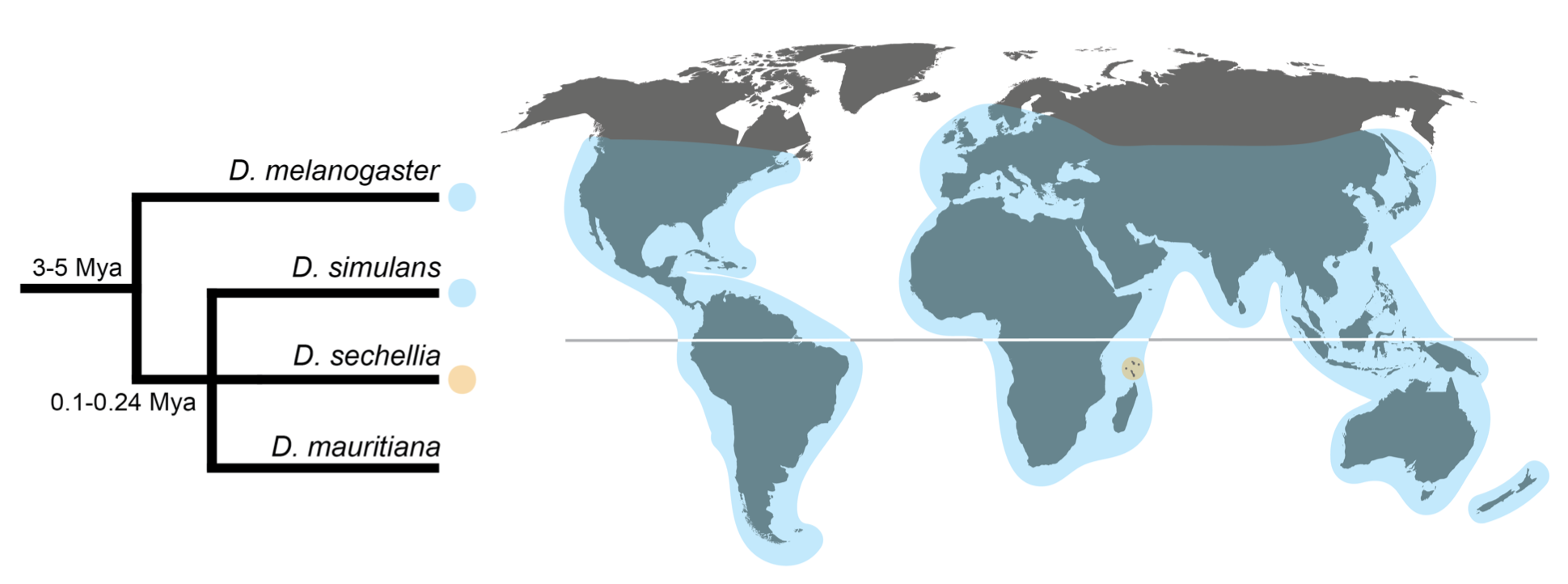
In many animals, circadian rhythms synchronize behavior with the timing of sunrise and sunset, coordinating specific behaviors with optimal activity periods throughout the day, avoiding environmental stressors, maximizing food availability, and aligning with fellow conspecifics for synchronized social/sexual behavior. However in most regions of the world, the timing of sunrise and sunset changes seasonally, and organisms are faced with days of variable lengths. Thus, circadian behaviors often display a high degree of phenotypic plasticity, allowing organisms to adjust the timing of their behaviors to better match with variable environments. The molecular and genetic basis of such behavioral plasticity remains largely unknown. Drosophilid flies generally display activity peaks surrounding sunrise and sunset, and have reduced movement throughout the day and night. I have identified differences in the degree of plasticity in the timing of these activity peaks for two Drosophila species: D. melanogaster, a highly-plastic, globally-distributed human commensal and D. sechellia, an equatorial island endemic that largely lacks circadian plasticity. My current work seeks to use these differences to test the hypothesis that changes in the molecular pacemaker mechanisms within circadian neurons drive differences in circadian behavioral plasticity, and circadian regulated behaviors. Read my most recent pre-print on this work here: https://www.biorxiv.org/content/10.1101/2023.07.05.547553v2
Past projects
The importance of male mate choice behavior
|
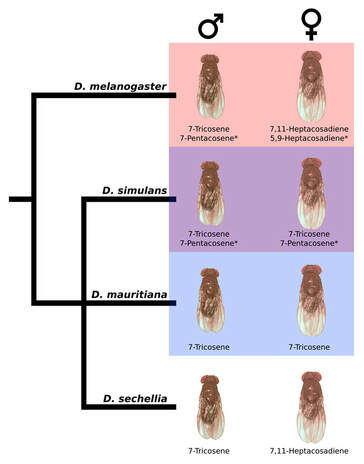
In Drosophila, pheromones act as important species identification signals that prevent mating between species. This work focuses on three species of Drosophila. In two of these species, D. melanogaster and D. sechellia, females express a blend of pheromones, the predominant one being 7,11-heptacosadiene (7,11-HD); D. simulans females express a different predominant pheromone, 7-Tricosene (7T). This is important, because 7,11-HD acts as a species identification signal. D. melanogaster and D. sechellia males readily court females that predominantly express 7,11-HD. However, for D. simulans, 7,11-HD is aversive and suppresses courtship behavior [1, 2]. My own work has shown that male mate discrimination in these species accounts for over 70% of gene flow limited by mating preferences, and may have evolved to alleviate the costs associated with heterospecific courtship and mating [2]. This work underscores the often overlooked importance of male mate choice in the accumulation of reproductive barriers.
|
The evolution of innate male mate choice behavior – signal receiver coevolution
The evolution of 7,11-HD expression is partially understood, and due in part to the evolution of two enzymes with large effect on CHC phenotype [3,4]. If the signal evolution is simple and likely rapid, how do receivers co-evolve to recognize a novel pheromone signal in time? I have harnessed the differences in male pheromone preference between these species, in combination with next generation DNA sequencing technology and transgenic fly strains, to map loci affecting male pheromone preference behavior in these species. Recent work has shown that the neural circuitry underlying male pheromone detection and response remains anatomically unchanged between D. simulans and D. melanogaster, and changes in preference are rather explained by a change in the interactions between neurons [5]. Identifying which genes underlie the evolved difference in pheromone preference behavior will provide valuable insight into the mechanism(s) underlying this change, however our results point to a complex genetic architecture involving multiple interacting loci making fine-mapping difficult [6].
The genetic architecture of an environmentally sensitive larval behavior
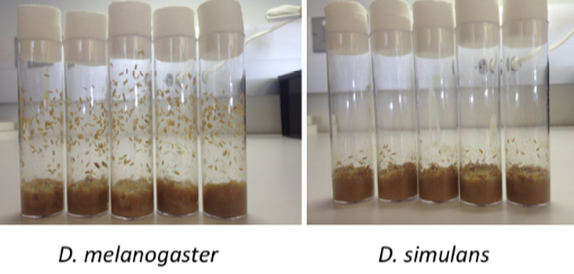
Some behaviors are difficult to study because they can be exquisitely sensitive to environmental variation. For example, before metamorphosis, Drosophila larvae enter a wandering stage, where they select a site to enter a non-mobile pupal stage (like a caterpillar's cocoon). Pupation site choice behavior in Drosophila can vary with larval density, substrate, humidity, temperature, and light condition. How then, is a simple insect larvae able to integrate so many stimuli to make a complex behavioral choice like selecting a pupation site? Despite this environmentally induced variation, the effects of genotype on preference are considerable, with the largest differences seen between species. Drosophila melanogaster, for instance, predominantly pupates away from the larval food source, while Drosophila simulans largely pupates on it [7]. We have used this difference in a highly controlled laboratory setting, in combination with deficiency mapping, RNAi knockdown, loss-of-function mutations, and qRT-PCR to collect evidence for two loci with substantial effects on pupation site choice preference [7]. Our results suggest that even complex behaviors may diverge via mutation of a small number of loci with sizable effects. Our results provide inroads to understanding the physiological mechanisms underlying the perception of the larval environment and the mechanisms regulating pupation site choice. Little is known about the selective forces driving pupation site choice behavioral evolution, but this work also provides a framework to begin studying the adaptive value of this behavior.
|
Literature cited:
[1] Billeter, J.-C., Atallah, J., Krupp, J. J., Millar, J. G., & Levine, J. D. (2009). Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature, 461(7266): 987–991. [2] Shahandeh, M. P., A. Pischedda, and T. L. Turner, (2018) Male mate choice via cuticular hydrocarbon pheromones drives reproductive isolation between Drosophila species. Evolution, 72: 123–135. [3] Shirangi TR, Dufour HD, Williams TM, Carroll SB. (2009). Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol 7(8):e1000168. [4] Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, et al. (2007). A female biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. PNAS, 104(11):4273–8. [5] Seeholzer LF, Seppo M, Stern DL, Ruta V., (2018) Evolution of a central neural circuit underlies Drosophila mate preferences. Nature, 559(7715):564–9. [6] Shahandeh, M.P. and Turner, T.L. (2020) Complex genetic architecture of male mate choice evolution between Drosophila species. Heredity, 124(6): 737-750. [7] A. Pischedda*, Shahandeh, M.P.*, Turner, T.L., (2020). The loci of behavioral evolution: evidence that Fas2 and tilB underlie differences in pupation site choice behavior between Drosophila melanogaster and D. simulans. Molecular Biology and Evolution, 37(3): 864-880. https://doi.org/10.1093/molbev/msz274 *denotes co-first authors |